ADNI Brain T1-Weighted MRI Images Classification and WGAN Generation (Alzheimer's and Healthy patients) for the purpose of data augmentation. Implemented in TensorFlow, trained on ADNI dataset.
- Abstract
- Report
- Requirements
- ADNI: Image Dataset Acquisition
- Image Preprocessing
- Wasserstein GAN Training and Generation
- Random and Stratified Undersampling
- Brain MRI Classification: Alzheimer's Disease (AD) and Cognitive Normal (CN) detection
- Status
- Contact
- License
- Contributing
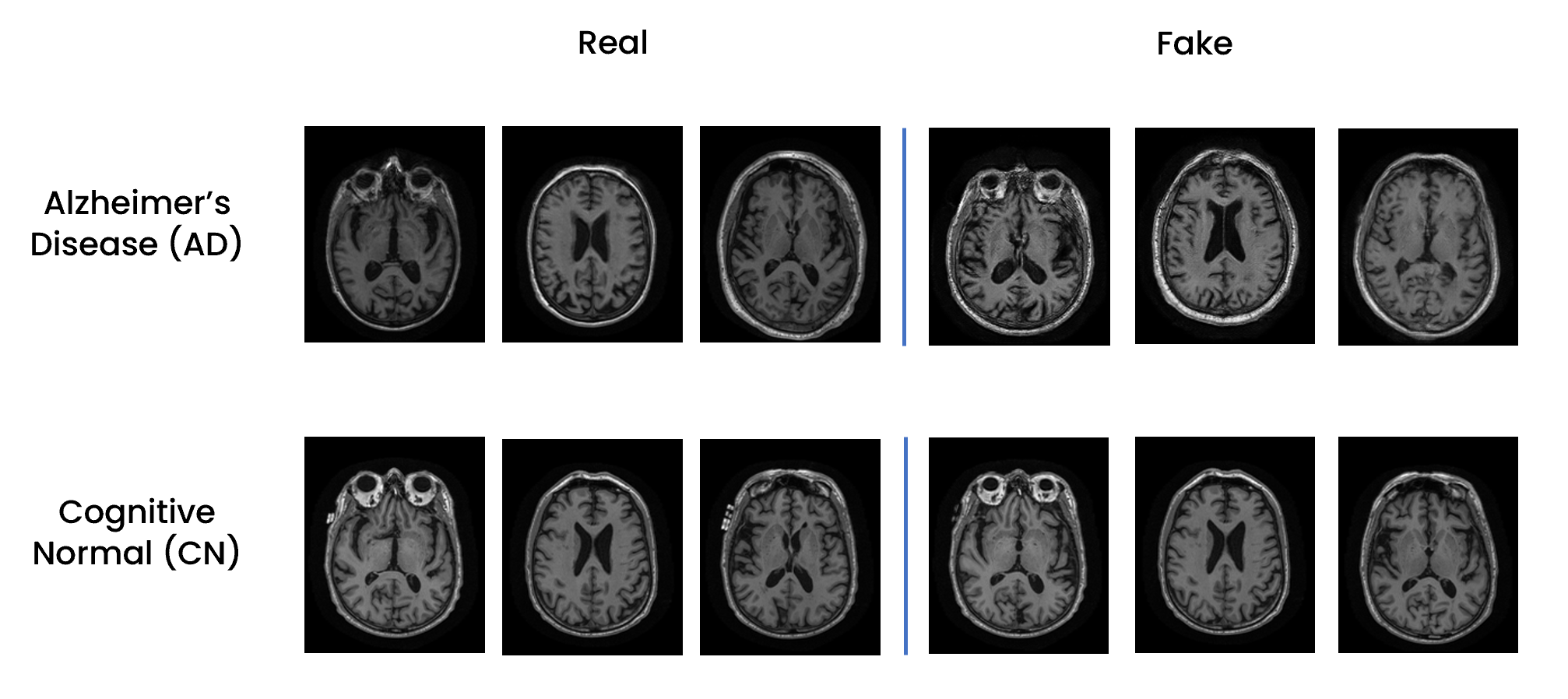
This project focused on Alzheimer's disease through three main objectives. Firstly, a dataset of axial 2D slices was created from 3D T1-weighted MRI brain images, integrating clinical, genetic, and biological sample data. Secondly, a Custom Resnet-18 was trained to classify these images, distinguishing between healthy individuals and those with Alzheimer's. The ultimate goal was to predict diagnoses solely based on cognitive assessments and clinical data. Lastly, different techniques for managing class imbalance were evaluated to improve the classifier's performance and reduce bias. Finally, a generative model (Wasserstein GAN) was trained on Alzheimer's Disease and Healthy for data augmentation and class balancing. This approach overcame the limitations of traditional undersampling, avoiding data loss and replacing problematic geometric data augmentation in the medical imaging context. The Wasserstein GAN provided an effective method to generate new samples of the minority class, contributing to improved performance of the final classifier.
- python==3.8
- ipython
- ipykernel
- matplotlib
- pandas
- seaborn
- tqdm==4.64.1
- scikit-learn==1.2.0
- glob2==0.7
- keras==2.10.0
- Keras-Preprocessing==1.1.2
- tensorflow==2.10.1
- numpy==1.24.2
- opencv_python==4.7.0.68
- pandas==1.5.3
- Pillow==9.4.0
- imageio==2.25.0
- Request access to the Alzheimer's Disease Neuroimaging Initiative: https://adni.loni.usc.edu/data-samples/access-data/ (tipically 1 week to be accepted)
- Visit the following website: https://ida.loni.usc.edu/login.jsp?project=ADNI&page=HOME#
- Sign up if you haven't already.
- Click on the "SEARCH" button.
- Select "Advanced Image Search (beta)."
- In the "Search Options" menu, choose "Pre-processed" under the "IMAGE TYPES" sub-menu.
- In the "Search Criteria" menu, select "MRI" under the "IMAGE" sub-menu. Then, under the "IMAGING PROTOCOL" sub-menu, choose "T1" for the weighting (Do not select "AXIAL" under the "Aquisition Plane" option, as most of the original data are volumetric .nii files.).
- To download subjects with Alzheimer's Disease (AD), go to the "Search Options" menu, select "Subject" under the "Search Sections" sub-menu, and then choose "AD" under the "subject" sub-menu. The same process applies to Normal Control (NC) subjects.
- Download only the "MPR; GradWarp; B1 Correction; N3; Scaled" images from the downloaded data.
- For compatibility, store the downloaded images of AD subjects in a directory named 'ad' and the images of NC subjects in a directory named 'nor'.
- Go in the
scripts/1_preprocessingfolder - Run the
1_ttv_split.pyscript to randomly split the patients into training, validation, and test sets. - Optionally, you can run the
2_subjects_lists.pyscript to log which subjects are assigned to each set. - Run the
3_nii_to_png_all.pyscript to convert the volumetric .nii data to axial .png MRI images. - Optionally, use the
4_check_shapes.ipynbnotebook to determine the number of different slices and dimensions for each visit's corresponding images. - Run the
5_select_sequences_ttv.pyscript to discard the images that are irrelevant for AD diagnosis, focusing only on the relevant parts of a subject's head. - Finally, execute the
6_resize_images.pyscript to resize all the images to a consistent size of (192, 160).
- Go in the
scripts/3_GAN/WGANfolder - Execute
wgan_bs_32_lat_128_eps_600.pyto train the GAN - Execute
generate_imgs.pyto generate the images
- Go to the
scripts/2_class_imbalance_managementfolder - Execute the various notebooks in it to perform class balancing
Use the notebooks in scripts/3_Resnet18_first folder to train and evaluate classification models for the detection of Alzheimer's Disease and Absence of symptoms in Brain MRI. Some forms of data augmentation are tested, including generation by trained W-GAN.
Giorgio Carbone - feel free to contact me!
-
You can check out the full license here
This project is licensed under the terms of the MIT license.
- Fork it (https://github.com/giocoal/ADNI-brain-MRI-alzheimer-WGAN-generation-and-classification.git)
- Create your feature branch (
git checkout -b feature/fooBar) - Commit your changes (
git commit -am 'Add some fooBar') - Push to the branch (
git push origin feature/fooBar) - Create a new Pull Request